The Human Genome Project, Gene Chips, Personalized Molecular Diagnostics, Bio-Cloud Computing... These hot words that attracted countless eyeballs in the first decade of the 21st century are all related to an industry – DNA sequencing. Biotechnology and information technology are blending in this new world of creativity. If you use a poem to describe the DNA sequencing industry that is escorted by two major technologies, it is that natural beauty is hard to give up.
With the completion of the Human Genome Project, humans have made unprecedented progress in understanding and mastering their own genetic information. At the same time, the molecular level detection technology platform has been continuously developed and improved, which has led to the rapid development of genetic detection technology and the continuous improvement of genetic detection efficiency. From the first generation of direct detection technology represented by Sanger sequencing and indirect sequencing technology represented by linkage analysis, by 2005, next-generation sequencing marked by Illumina's Solexa technology and ABI's SOLiD technology (next- Generation sequencing (NGS) has emerged, the sequencing efficiency has been significantly improved, the time has been significantly shortened, the cost has been significantly reduced, and the genetic testing methods have revolutionized.
DNA sequencing technology has undergone different stages of development. This article will focus on the basic principles of first-, second-, and third-generation sequencing, representative sequencing platforms, and the characteristics of sequencing technologies at different stages of development.
First generation sequencing technology
Traditional dideoxy chain termination methods, chemical degradation methods, and various DNA sequencing technologies developed on the basis of these are collectively referred to as first generation DNA sequencing technologies. The human genome project (HGP) is based primarily on first-generation DNA sequencing technology.
Dideoxy chain termination
The dideoxy chain termination method is also known as the Sanger method. The principle of the method is: when the nucleic acid template is replicated in the presence of DNA polymerase, primers, and four monodeoxynucleoside triphosphates (dNTPs, one of which is labeled with radioactive P32), the ratio is proportional to the four-tube reaction system. Introduced 4 dideoxynucleoside triphosphates (ddNTPs), because dideoxynucleosides do not have 3'OH, so as long as the dideoxynucleosides are incorporated into the end of the chain, the chain stops prolonging, if the chain ends are doped with monodeoxynucleosides The chain can continue to be extended. Thus, a series of nucleic acid fragments of different lengths each having a 3' end of the dideoxy base are synthesized in each reaction system. After the reaction was terminated, gel electrophoresis was carried out in four lanes to separate nucleic acid fragments of different lengths, and adjacent fragments of one length differ by one base. After autoradiography, the base sequence of the synthetic fragments can be read sequentially according to the dideoxynucleotides at the 3' end of the fragment.
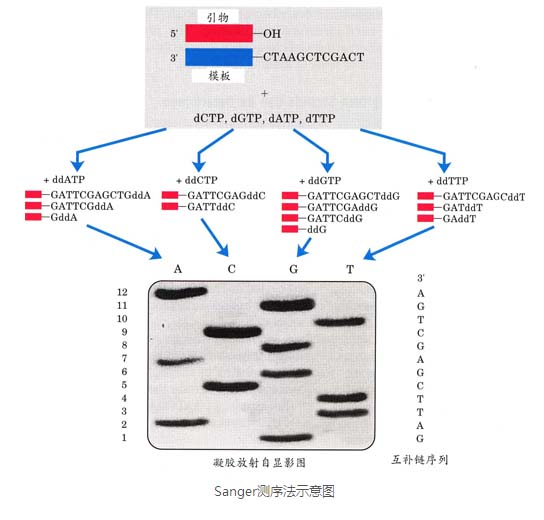
The main features of the first generation sequencing technology are:
- The sequencing read length can reach 1000bp;
- Accuracy is as high as 99.999%;
- High cost of sequencing;
- Low flux.
Second Generation Sequencing Technology (NGS)
With the completion of the Human Genome Project, people entered the post-genome era, the functional genomic era, and traditional sequencing methods have been unable to meet the needs of large-scale genome sequencing such as deep sequencing and re-sequencing, which has led to the birth of a new generation of DNA sequencing technology. . Next-generation sequencing technology, also known as second-generation sequencing technology, includes Roche 454's GS FLX sequencing platform, Illumina's Solexa Genome Analyzer sequencing platform, and ABI's SOLID sequencing platform.
The core idea generation sequencing is sequencing-by- synthesis (Sequencing by Synthesis), i.e. to determine the sequence of the DNA marker by capturing the end of the newly synthesized. Take the technical principle of Illumina as an example: firstly, the surface of the fragment DNA is connected to the surface of the fragment, and the purpose is to match the chip on the chip, then hybridize with the chip, bridge amplification, and then the formation of the cluster is to ensure The signal intensity, followed by the addition of four blockers and fluorescently labeled single nucleotides, is cycled, recording the fluorescence information for each cycle, and finally obtaining the sequence information.
The most striking feature of second-generation sequencing technology is the high-throughput sequence of sequencing hundreds of thousands to millions of DNA molecules at a time, making it easy to sequence transcriptomes or deep genome sequencing of a species. The cost has also dropped significantly.
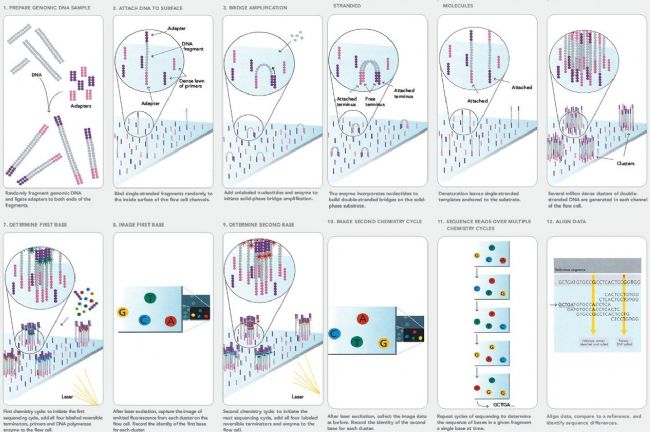
Illumina sequencing process
Third generation sequencing technology
In recent years, sequencing technology has made new progress and developed into third-generation sequencing. The core concept of the third-generation sequencing technology is to synthesize the side synthesis target with single molecule. This technique does not require PCR amplification, enabling separate sequencing of each DNA molecule. Representative sequencing platforms are PacBio's SMRT and Oxford Nanopore Technologies nanopore single molecule sequencing technology.
Among them, PacBio SMRT technology also applies the idea of ​​sequencing while synthesizing, and uses SMRT chip as the sequencing carrier. The basic principle is: DNA polymerase and template are combined, 4 colors are fluorescently labeled with 4 bases (ie, dNTP). In the base pairing stage, different bases are added, and different light is emitted, which can be judged according to the wavelength and peak value of light. The type of base entered. At the same time, this DNA polymerase is one of the keys to achieving ultra-long read length. The sequencing of SMRT technology is fast, with about 10 dNTPs per second.
The third-generation sequencing equipment is superior to the second-generation equipment in the length of the DNA sequence fragment, but the accuracy is worse than that of the second-generation equipment. In the future, with the improvement of technology, the third-generation sequencing equipment will be more stable and mature.
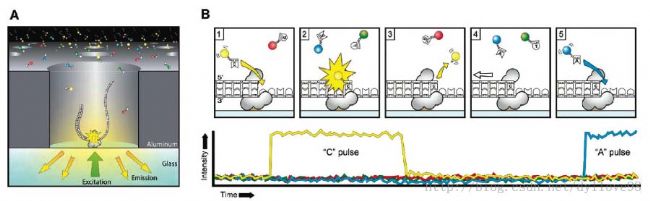
PacBio SMRT sequencing principle
After more than 30 years of development, DNA sequencing technology has reached the third generation, and the three-generation sequencing technology has its own advantages. In the future, based on the purpose of different sequencing, the application of three generations of sequencing technology will each play its role, and the cost of sequencing will be further reduced. The rapid development of sequencing technology will also promote the progress of biological research and medical applications.
Microscope for Biology,Bio Lab Microscope,Professional Biological Microscope,Microscopes For Biology
Ningbo ProWay Optics & Electronics Co., Ltd. , https://www.proway-microtech.com